|
MHC class I
MHC class I molecules are one of two primary classes of major histocompatibility complex (MHC) molecules (the other being MHC class II) and are found on the cell surface of all nucleated cells in the bodies of vertebrates.[1][2] They also occur on platelets, but not on red blood cells. Their function is to display peptide fragments of proteins from within the cell to cytotoxic T cells; this will trigger an immediate response from the immune system against a particular non-self antigen displayed with the help of an MHC class I protein. Because MHC class I molecules present peptides derived from cytosolic proteins, the pathway of MHC class I presentation is often called cytosolic or endogenous pathway.[3] In humans, the HLAs corresponding to MHC class I are HLA-A, HLA-B, and HLA-C. FunctionClass I MHC molecules bind peptides generated mainly from the degradation of cytosolic proteins by the proteasome. The MHC I: peptide complex is then inserted via the endoplasmic reticulum into the external plasma membrane of the cell. The epitope peptide is bound on extracellular parts of the class I MHC molecule. Thus, the function of the class I MHC is to display intracellular proteins to cytotoxic T cells (CTLs). However, class I MHC can also present peptides generated from exogenous proteins, in a process known as cross-presentation. A normal cell will display peptides from normal cellular protein turnover on its class I MHC, and CTLs will not be activated in response to them due to central and peripheral tolerance mechanisms. When a cell expresses foreign proteins, such as after viral infection, a fraction of the class I MHC will display these peptides on the cell surface. Consequently, CTLs specific for the MHC:peptide complex will recognize and kill presenting cells. Alternatively, class I MHC itself can serve as an inhibitory ligand for natural killer cells (NKs). Reduction in the normal levels of surface class I MHC, a mechanism employed by some viruses[4] and certain tumors to evade CTL responses, activates NK cell killing. PirB and visual plasticityPaired-immunoglobulin-like receptor B (PirB), an MHCI-binding receptor, is involved in the regulation of visual plasticity.[5] PirB is expressed in the central nervous system and diminishes ocular dominance plasticity in the developmental critical period and adulthood.[5] When the function of PirB was abolished in mutant mice, ocular dominance plasticity became more pronounced at all ages.[5] PirB loss of function mutant mice also exhibited enhanced plasticity after monocular deprivation during the critical period.[5] These results suggest that PirB may be involved in the modulation of synaptic plasticity in the visual cortex. StructureMHC class I molecules are heterodimers that consist of two polypeptide chains, α and β2-microglobulin (B2M). The two chains are linked noncovalently via interaction of B2M and the α3 domain. Only the α chain is polymorphic and encoded by a HLA gene, while the B2M subunit is not polymorphic and encoded by the Beta-2 microglobulin gene. The α3 domain is plasma membrane-spanning and interacts with the CD8 co-receptor of T-cells. The α3-CD8 interaction holds the MHC I molecule in place while the T cell receptor (TCR) on the surface of the cytotoxic T cell binds its α1-α2 heterodimer ligand, and checks the coupled peptide for antigenicity. The α1 and α2 domains fold to make up a groove for peptides to bind. MHC class I molecules bind peptides that are predominantly 8-10 amino acid in length (Parham 87), but the binding of longer peptides have also been reported.[6] While a high-affinity peptide and the B2M subunit are normally required to maintain a stable ternary complex between the peptide, MHC I, and B2M, under subphysiological temperatures, stable, peptide-deficient MHC I/B2M heterodimers have been observed.[7][8] Synthetic stable, peptide-receptive MHC I molecules have been generated using a disulfide bond between the MHC I and B2M, named "open MHC-I".[9] Synthesis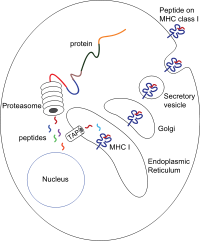 The peptides are generated mainly in the cytosol by the proteasome. The proteasome is a macromolecule that consists of 28 subunits, of which half affect proteolytic activity. The proteasome degrades intracellular proteins into small peptides that are then released into the cytosol. Proteasomes can also ligate distinct peptide fragments (termed spliced peptides), producing sequences that are noncontiguous and therefore not linearly templated in the genome. The origin of spliced peptide segments can be from the same protein (cis-splicing) or different proteins (trans-splicing).[10][11] The peptides have to be translocated from the cytosol into the endoplasmic reticulum (ER) to meet the MHC class I molecule, whose peptide-binding site is in the lumen of the ER. They have membrane proximal Ig fold Translocation and peptide loadingThe peptide translocation from the cytosol into the lumen of the ER is accomplished by the transporter associated with antigen processing (TAP). TAP is a member of the ABC transporter family and is a heterodimeric multimembrane-spanning polypeptide consisting of TAP1 and TAP2. The two subunits form a peptide binding site and two ATP binding sites that face the cytosol. TAP binds peptides on the cytoplasmic side and translocates them under ATP consumption into the lumen of the ER. The MHC class I molecule is then, in turn, loaded with peptides in the lumen of the ER. The peptide-loading process involves several other molecules that form a large multimeric complex called the Peptide loading complex[12] consisting of TAP, tapasin, calreticulin, calnexin, and Erp57 (PDIA3). Calnexin acts to stabilize the class I MHC α chains prior to β2m binding. Following complete assembly of the MHC molecule, calnexin dissociates. The MHC molecule lacking a bound peptide is inherently unstable and requires the binding of the chaperones calreticulin and Erp57. Additionally, tapasin binds to the MHC molecule and serves to link it to the TAP proteins and facilitates the selection of peptide in an iterative process called peptide editing,[13][14][15] thus facilitating enhanced peptide loading and colocalization. Once the peptide is loaded onto the MHC class I molecule, the complex dissociates and it leaves the ER through the secretory pathway to reach the cell surface. The transport of the MHC class I molecules through the secretory pathway involves several posttranslational modifications of the MHC molecule. Some of the posttranslational modifications occur in the ER and involve change to the N-glycan regions of the protein, followed by extensive changes to the N-glycans in the Golgi apparatus. The N-glycans mature fully before they reach the cell surface. Peptide removalPeptides that fail to bind MHC class I molecules in the lumen of the endoplasmic reticulum (ER) are removed from the ER via the sec61 channel into the cytosol,[16][17] where they might undergo further trimming in size, and might be translocated by TAP back into ER for binding to a MHC class I molecule. For example, an interaction of sec61 with bovine albumin has been observed.[18] Effect of virusesMHC class I molecules are loaded with peptides generated from the degradation of ubiquitinated cytosolic proteins in proteasomes. As viruses induce cellular expression of viral proteins, some of these products are tagged for degradation, with the resulting peptide fragments entering the endoplasmic reticulum and binding to MHC I molecules. It is in this way, the MHC class I-dependent pathway of antigen presentation, that the virus infected cells signal T-cells that abnormal proteins are being produced as a result of infection. The fate of the virus-infected cell is almost always induction of apoptosis through cell-mediated immunity, reducing the risk of infecting neighboring cells. As an evolutionary response to this method of immune surveillance, many viruses are able to down-regulate or otherwise prevent the presentation of MHC class I molecules on the cell surface. In contrast to cytotoxic T lymphocytes, natural killer (NK) cells are normally inactivated upon recognizing MHC I molecules on the surface of cells. Therefore, in the absence of MHC I molecules, NK cells are activated and recognize the cell as aberrant, suggesting that it may be infected by viruses attempting to evade immune destruction. Several human cancers also show down-regulation of MHC I, giving transformed cells the same survival advantage of being able to avoid normal immune surveillance designed to destroy any infected or transformed cells.[19] Genes and isotypes
Evolutionary historyThe MHC class I genes originated in the most recent common ancestor of all jawed vertebrates, and have been found in all living jawed vertebrates that have been studied thus far.[2] Since their emergence in jawed vertebrates, this gene family has been subjected to many divergent evolutionary paths as speciation events have taken place. There are, however, documented cases of trans-species polymorphisms in MHC class I genes, where a particular allele in an evolutionary related MHC class I gene remains in two species, likely due to strong pathogen-mediated balancing selection by pathogens that can infect both species.[20] Birth-and-death evolution is one of the mechanistic explanations for the size of the MHC class I gene family. Birth-and-death of MHC class I genesBirth-and-death evolution asserts that gene duplication events cause the genome to contain multiple copies of a gene which can then undergo separate evolutionary processes. Sometimes these processes result in pseudogenization (death) of one copy of the gene, though sometimes this process results in two new genes with divergent function.[21] It is likely that human MHC class Ib loci (HLA-E, -F, and -G) as well as MHC class I pseudogenes arose from MHC class Ia loci (HLA-A, -B, and -C) in this birth-and-death process.[22] References
External links
|
||||||||||
